In the proviral pipeline, two main tasks are performed with regard to primers. First, the pipeline verifies that the correct primer sequences appear at the ends of each sample. Second, after confirming these primers are present, the pipeline removes (or “strips”) them so that only the viral genomic data remain for further analysis. These steps help ensure that downstream results accurately reflect the virus rather than any extra sequences added during laboratory processing.
What Primers Are and Why They Matter
- Primers are short DNA sequences that are used during PCR amplification to target a specific region of the viral genome.
- For the proviral pipeline, only one set of primers is supported. The forward primer is defined as
GCGCCCGAACAGGGACYTGAAARCGAAAG(and a “no-mixture” variant is available to remove any ambiguous letters), and the reverse primer isTAAGCCTCAATAAAGCTTGCCTTGAGTGC. - These specific primers tell the pipeline where the viral sequence begins and ends. However, once these locations are defined, it’s important to remove the primer sequences before running further analyses so that the extra nonviral bits do not distort downstream results.
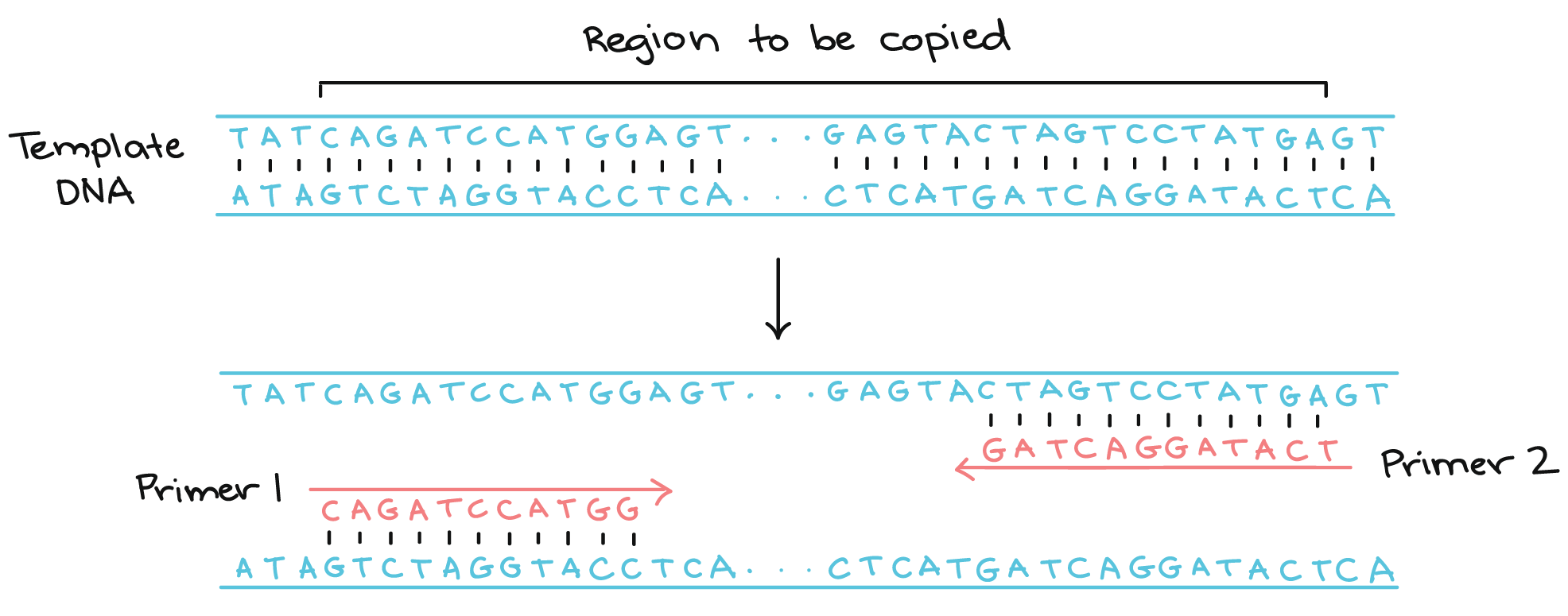
How the Pipeline Handles Primers
- Quality Check and Primer Search
- Before searching for primers, the system verifies that each sample has strong, high–coverage sequence data (for example, that there aren’t too many unclear bases). It also skips samples that are not expected to be proviral.
- The pipeline then examines each sample’s beginning (the 5’ end) and end (the 3’ end) using a preset “probe” length. It looks for the exact primer sequences noted above.
- If a primer isn’t immediately detected, the pipeline also checks whether the sample might be in the reverse orientation (by “flipping” the sequence). If the flipped version shows the primers in the right positions, it will use that result and flag the sample accordingly.
- Removing the Primers
- Once at least one valid primer has been found at each end, the pipeline “strips” these primer sequences off.
- Removing the primers is crucial because the sequences added during the PCR process aren’t part of the virus itself. Keeping them might lead to inaccuracies when looking for genuine viral mutations or defects.
- Error Reporting and User Guidance
- If the pipeline cannot find a primer, or if the match is too short or otherwise doesn’t meet quality standards, an error is recorded. Common error messages include
primer was not foundorprimer failed validation. - A special flag (
is_rev_comp) is also added if the system had to flip the sequence to detect valid primers. This information is provided in the output so you can review any unexpected issues.
- If the pipeline cannot find a primer, or if the match is too short or otherwise doesn’t meet quality standards, an error is recorded. Common error messages include
Tips for Users
- If errors occur in primer detection or removal, you’ll see clear messages in the output. These messages indicate whether you should double-check your laboratory protocols — for example, whether the correct primer set was used — or take other troubleshooting steps.
- Errors such as
low end read coveragesuggest that a portion of the sequence might have been trimmed during quality filtering. This may happen if the input sequence is low quality. - A reverse complement flag (
is_rev_comp) in the output means that one or both primers were only detected when the sequence was flipped. In most cases, this is not a problem but can help explain unexpected results.
Summary
The pipeline first checks each sample for quality and then searches both ends for the expected primer sequences. Once found, these primer sequences are removed from the sample so that only the genuine proviral DNA is analyzed. This two-step process ensures that your final results reflect only the viral genome without any artifacts from the amplification process. If a sample fails these checks, the pipeline provides easy-to-understand error messages so that you know which ones may need further attention.